FCM Data Clustering
Description
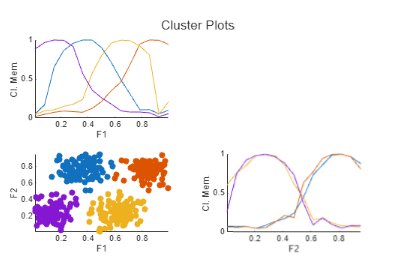
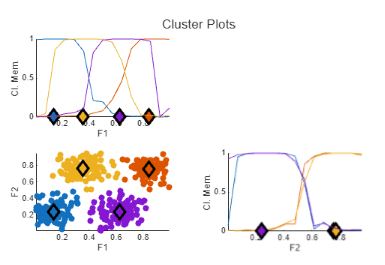
The FCM Data Clustering task clusters data using the fuzzy c-means (FCM) algorithm, where each data point belongs to a cluster to a degree that is specified by a membership grade. For example, a data point that lies close to the center of a cluster will have a high degree of membership in that cluster, and another data point that lies far away from the center of a cluster will have a low degree of membership to that cluster. The FCM Data Clustering task automatically generates MATLAB® code for your live script. For more information about Live Editor tasks, see Add Interactive Tasks to a Live Script.
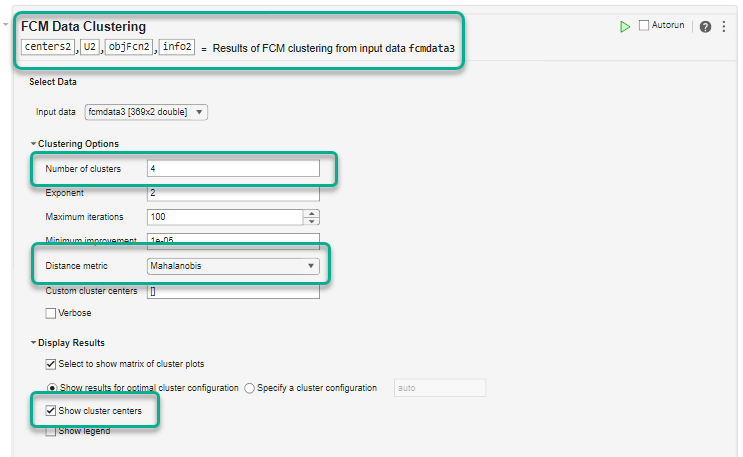
The task returns these output arguments from the fcm
function:
centers— Cluster centersU— Fuzzy partition matrix indicating the degree of membership of each data point in each clusterobjFcn— Objective function values for each clustering iterationinfo— Detailed clustering results
For more information on the FCM algorithm, see Fuzzy Clustering.
Open the Task
To add the FCM Data Clustering task to a live script in the MATLAB Editor:
On the Live Editor tab, select Task > FCM Data Clustering.
In a code block in the script, enter a relevant keyword, such as
fcmorclustering. Select FCM Data Clustering from the suggested command completions.
Parameters
Version History
Introduced in R2025a