connect
Syntax
Description
connect(
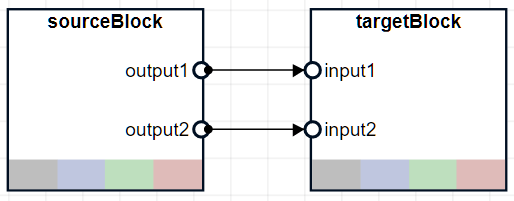
connects an output port of the pipeline,sourceBlock,targetBlock,portsToConnect)sourceBlock to an input port of the
targetBlock. portsToConnect specifies the output
and input ports.
mappedPorts = connect(pipeline,sourceBlock,targetBlock,portsToConnect)mappedPorts between the
source block and target block after connecting two blocks as specified by
portsToConnect.
Examples
Input Arguments
Output Arguments
Version History
Introduced in R2023a