mzcdfread
Read mass spectrometry data from netCDF file
Syntax
mzCDFStruct = mzcdfread(File)
mzCDFStruct = mzcdfread(File,
...'TimeRange', TimeRangeValue, ...)
mzCDFStruct = mzcdfread(File,
...'ScanIndices', ScanIndicesValue, ...)
mzCDFStruct = mzcdfread(File,
...'Verbose', VerboseValue, ...)
Input Arguments
File | Character vector or string containing a file name, or a path and file name, of a netCDF file that contains mass spectrometry data and conforms to the ANDI/MS or the ASTM E2077-00 (2005) standard specification or earlier specifications. If you specify only a file name, that file must be on the MATLAB® search path or in the current folder. |
TimeRangeValue | Two-element numeric array Tip Time units are indicated in the netCDF global attributes. For
summary information about the time ranges in a netCDF file, use the Note If you specify a |
ScanIndicesValue | Positive integer, vector of integers, or a two-element
numeric array Tip For information about the scan indices in a netCDF file, check
the Note If you specify a |
VerboseValue | Controls the display of the progress of the reading of |
Output Arguments
mzCDFStruct | MATLAB structure containing mass spectrometry information
from a netCDF file. Its fields correspond to the variables and global
attributes in a netCDF file. If a netCDF variable contains local attributes,
an additional field is created, with the name of the field being the
variable name appended with |
Description
mzCDFStruct = mzcdfread(File)File, and then creates a MATLAB structure, mzCDFStruct.
File is a character vector or string containing a file name, or a
path and file name, of a netCDF file that contains mass spectrometry data. The file must
conform to the ANDI/MS or the ASTM E2077-00 (2005) standard specification or earlier
specifications.
mzCDFStruct contains fields that
correspond to the variables and global attributes in a netCDF file.
If a netCDF variable contains local attributes, an additional field
is created, with the name of the field being the variable name appended
with _attributes. The number and names of the fields
will vary, depending on the mass spectrometer software, but typically
there are mass_values and intensity_values fields.
Tip
LC/MS data analysis requires extended amounts of memory from the operating system.
If you receive errors related to memory, try the following:
Increase the virtual memory (swap space) for your operating system as described in Resolve “Out of Memory” Errors.
If you receive errors related to Java® heap space, increase your Java heap space:
If you have MATLAB version 7.10 (R2010a) or later, see Java Heap Memory Settings.
If you have MATLAB version 7.9 (R2009b) or earlier, see https://www.mathworks.com/matlabcentral/answers/92813-how-do-i-increase-the-heap-space-for-the-java-vm-in-matlab.
mzCDFStruct = mzcdfread(File,
...'PropertyName', PropertyValue,
...)mzcdfread with optional properties
that use property name/property value pairs. You can specify one or
more properties in any order. Each PropertyName must
be enclosed in single quotation marks and is case insensitive. These
property name/property value pairs are as follows:
specifies
the range of time in mzCDFStruct = mzcdfread(File,
...'TimeRange', TimeRangeValue, ...)File to read. TimeRangeValue is
a two-element numeric array [Start End]. Default
is to read spectra from all times [0 Inf].
Tip
Time units are indicated in the netCDF global attributes. For
summary information about the time ranges in a netCDF file, use the mzcdfinfo function.
Note
If you specify a TimeRangeValue,
you cannot specify ScanIndicesValue.
mzCDFStruct = mzcdfread(File,
...'ScanIndices', ScanIndicesValue, ...)File to
read. ScanIndicesValue is a positive integer,
vector of integers, or a two-element numeric array [Start_Ind
End_Ind]. Start_Ind and End_Ind are
each positive integers indicating a scan index number. Start_Ind must
be less than End_Ind. Default is to read all scans.
Tip
For information about the scan indices in a netCDF file, check
the NumberOfScans field in the structure returned
by the mzcdfinfo function.
Note
If you specify a ScanIndicesValue,
you cannot specify a TimeRangeValue.
mzCDFStruct = mzcdfread(File,
...'Verbose', VerboseValue, ...)File.
Choices are true (default) or false.
Examples
In the following example, the file results.cdf is
not provided.
Read a netCDF file into the MATLAB software as a structure.
out = mzcdfread('results.cdf');View the second scan in the netCDF file by creating separate variables containing the intensity and m/z values, and then plotting these values. Add a title and x- and y-axis labels using fields in the output structure.
idx1 = out.scan_index(2)+1; idx2 = out.scan_index(3); y = out.intensity_values(idx1:idx2); z = out.mass_values(idx1:idx2); stem(z,y,'marker','none') title(sprintf('Time: %f',out.scan_acquisition_time(2))) xlabel(out.mass_axis_units) ylabel(out.intensity_axis_units)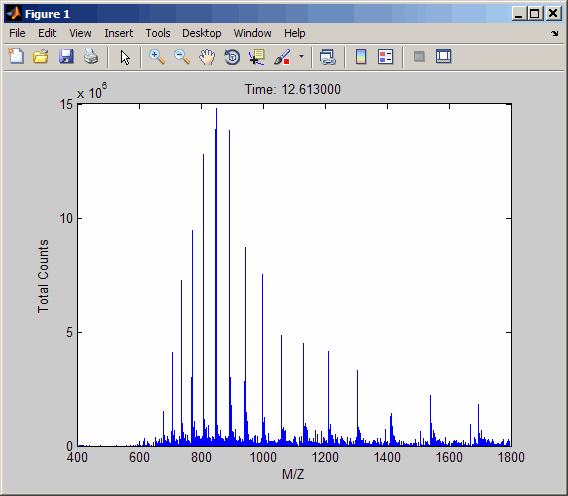
Version History
Introduced in R2008b
See Also
jcampread | mzcdf2peaks | mzcdfinfo | mzxmlread | tgspcread