JuGEx - Julich-Brain Gene Expression
JuGEx - Julich-Brain Gene Expression
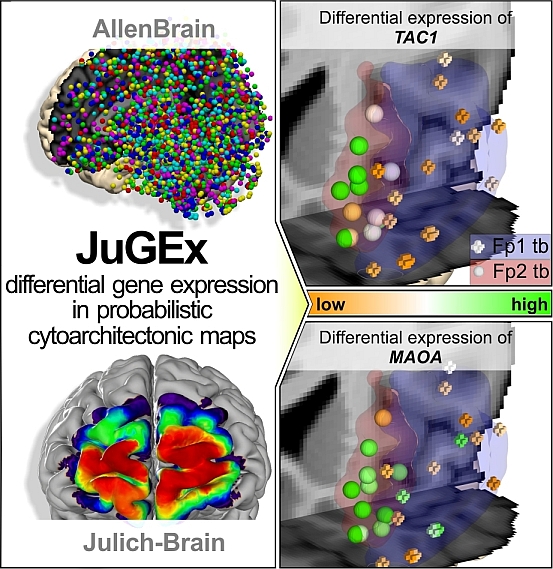
Cytoarchitecture linked to gene expression to study multilevel human brain organization
Decoding the chain from genes to cognition requires detailed insights how areas with specific gene activities and microanatomical architectures contribute to brain function and dysfunction. The Allen Human Brain Atlas [1] contains regional gene expression data, while the Julich-Brain Atlas [2] offers three-dimensional cytoarchitectonic maps reflecting the interindividual variability. JuGEx offers an integrated framework that combines the analytical benefits of both repositories towards a multi-level brain atlas of adult humans. JuGEx is a new method for integrating tissue transcriptome and cytoarchitectonic segregation.
JuGEx finds differentially expressed genes based on a user-defined candidate gene list between two user-defined volumes of interest (Julich-Brain maps or other volume of interest in MNI152 reference space [3]). The tool downloads expression values via the Allen Brain API, which are further analysed using a permuted n-way ANOVA.
Important: Make sure that your VOIs are registered to the MNI152 reference space! If not, you will extract wrong and non-corresponding tissue samples through the Allen Brain API.
- [1] Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL et al (2012) An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489:391–399. https://doi.org/10.1038/nature11405
- [2] Amunts, K., Mohlberg, H., Bludau, S., Zilles, K. (2020). Julich-Brain – A 3D probabilistic atlas of human brain’s cytoarchitecture. Science 369, 988-992. DOI: 10.1126/science.abb4588
- [3] Evans, A. C., Janke, A. L., Collins, D. L., Baillet, S. (2012). Brain templates and atlases. Neuroimage, 62(2):911-22. DOI: 10.1016/j.neuroimage.2012.01.024
Cite As
Bludau, Sebastian, et al. “Integration of Transcriptomic and Cytoarchitectonic Data Implicates a Role for MAOA and TAC1 in the Limbic-Cortical Network.” Brain Structure and Function, vol. 223, no. 5, Springer Science and Business Media LLC, Feb. 2018, pp. 2335–42, doi:10.1007/s00429-018-1620-6.
Cita come
Bludau, Sebastian, et al. “Integration of Transcriptomic and Cytoarchitectonic Data Implicates a Role for MAOA and TAC1 in the Limbic-Cortical Network.” Brain Structure and Function, vol. 223, no. 5, Springer Science and Business Media LLC, Feb. 2018, pp. 2335–42, doi:10.1007/s00429-018-1620-6.
Compatibilità della release di MATLAB
Compatibilità della piattaforma
Windows macOS LinuxCategorie
Tag
Riconoscimenti
Ispirato da: superSlider, xml2struct, Arrow3, BUBBLEPLOT3, Winsorize data
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Scopri Live Editor
Crea script con codice, output e testo formattato in un unico documento eseguibile.
JuGex
JuGex/analyze_AB_exp_Data
JuGex/extract_AB_exp_Data
JuGex/utility
JuGex/visualize_AB_exp_Data
| Versione | Pubblicato | Note della release | |
|---|---|---|---|
| 1.0.2 | See release notes for this release on GitHub: https://github.com/INM-1-FZJ-Cytoarchitecture/jugex/releases/tag/1.0.2 |
||
| 1.0.0 |
|