updateOrientation
Syntax
Description
medVolReoriented = updateOrientation(medVol,direction)medicalVolume
object medVol to the specified direction,
direction. The output volume contains the reoriented voxel data in
its Voxels property, and other property values update to reflect the
intrinsic-to-world mapping between the new voxel data and the patient coordinate system. The
position of the volume in patient coordinates remains unchanged. This syntax applies default
conventions for the order and positive direction of each dimension of the voxel data.
medVolReoriented = updateOrientation(medVol,orientationCode)medVol using the order and positive
direction for each dimension of the voxel data specified by an orientation code.
Examples
Update the primary slice orientation of a CT volume from transverse to coronal.
Download Images
Download a file containing two CT chest volumes from the Medical Segmentation Decathlon data set [1]. The size of the data file is approximately 76 MB. Run this code to download the data set from the MathWorks® website and unzip the folder.
zipFile = matlab.internal.examples.downloadSupportFile("medical","MedicalVolumeNIfTIData.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath) dataFolder = fullfile(filepath,"MedicalVolumeNIfTIData");
Specify the path of the folder that contains the downloaded and unzipped data for the first volume.
filePath = fullfile(dataFolder,"lung_027.nii.gz");Import Data as Medical Volume
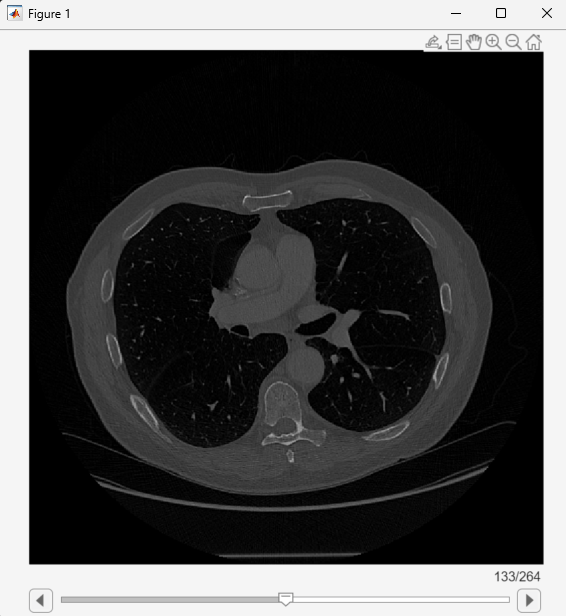
Load the CT volume as a medicalVolume object. Note that the Orientation property value is "transverse", indicating the primary orientation is the transverse plane.
medVol = medicalVolume(filePath)
medVol =
medicalVolume with properties:
Voxels: [512×512×264 single]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "mm"
Orientation: "transverse"
VoxelSpacing: [0.8594 0.8594 1.2453]
NormalVector: [0 0 -1]
NumCoronalSlices: 512
NumSagittalSlices: 512
NumTransverseSlices: 264
PlaneMapping: ["sagittal" "coronal" "transverse"]
DataDimensionMeaning: ["left" "anterior" "superior"]
Modality: "unknown"
WindowCenters: 0
WindowWidths: 0
View the medicalVolume object using sliceViewer, which opens a new scrollable window that displays transverse slices.
sliceViewer(medVol)
Update Orientation to Coronal
Update the primary orientation to the coronal plane.
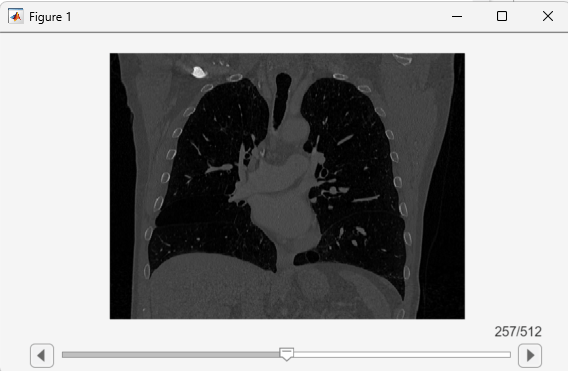
In the updated medicalVolume, the Orientation property value is "coronal". The DataDimensionMeaning property indicates the order and positive direction for all three dimensions of the voxel data in the Voxels property. When you specify the new orientation as a scalar string, the function uses default directions for the other dimensions. To fully define the new orientation, specify the orientationCode input argument as a 3-element string array.
medVolReoriented = updateOrientation(medVol,"coronal")medVolReoriented =
medicalVolume with properties:
Voxels: [264×512×512 single]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "unknown"
Orientation: "coronal"
VoxelSpacing: [0.8594 1.2453 0.8594]
NormalVector: [0 1 0]
NumCoronalSlices: 512
NumSagittalSlices: 512
NumTransverseSlices: 264
PlaneMapping: ["transverse" "sagittal" "coronal"]
DataDimensionMeaning: ["inferior" "left" "posterior"]
Modality: "unknown"
WindowCenters: []
WindowWidths: []
View the updated volume using sliceViewer, verifying that the window displays coronal slices.
sliceViewer(medVolReoriented)
[1] Medical Segmentation Decathlon. "Lung." Tasks. Accessed May 10, 2018. http://medicaldecathlon.com/.
The Medical Segmentation Decathlon data set is provided under the CC-BY-SA 4.0 license. All warranties and representations are disclaimed. See the license for details.
Update the orientation of a CT volume, specifying the positive direction of each dimension by using the orientationCode input argument. Use this approach when you require the voxel data in a specific, non-default orientation, such as when preprocessing data for a deep learning network with an expected input orientation.
Download Image Data
Download a file containing two CT chest volumes from the Medical Segmentation Decathlon data set [1]. The size of the data file is approximately 76 MB. Run this code to download the data set from the MathWorks® website and unzip the folder.
zipFile = matlab.internal.examples.downloadSupportFile("medical","MedicalVolumeNIfTIData.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath) dataFolder = fullfile(filepath,"MedicalVolumeNIfTIData");
Specify the path of the folder that contains the downloaded and unzipped data for the first volume.
filePath = fullfile(dataFolder,"lung_027.nii.gz");Import Data as Medical Volume
Load the CT volume as a medicalVolume object.
The Orientation property value indicates the primary orientation is the transverse plane. The DataDimensionMeaning property value indicates that the row indices of the voxel data increase in the left direction, the column indices increase in the anterior direction, and the slice indices increase in the superior direction.
medVol = medicalVolume(filePath)
medVol =
medicalVolume with properties:
Voxels: [512×512×264 single]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "mm"
Orientation: "transverse"
VoxelSpacing: [0.8594 0.8594 1.2453]
NormalVector: [0 0 -1]
NumCoronalSlices: 512
NumSagittalSlices: 512
NumTransverseSlices: 264
PlaneMapping: ["sagittal" "coronal" "transverse"]
DataDimensionMeaning: ["left" "anterior" "superior"]
Modality: "unknown"
WindowCenters: 0
WindowWidths: 0
Update Orientation
Orient the volume such that the row indices increase in the posterior direction, column indices increase in the right direction, and slice indices increase in the inferior direction. Specify the target orientation as a three-element string array, where each string specifies the positive anatomical direction for the corresponding voxel dimension.
In the updated volume, the Orientation property value is still "transverse", because the primary direction remains unchanged. However, the DataDimensionMeaning property value indicates that the directions and the order of the first two dimensions have updated.
medVolReoriented = updateOrientation(medVol,["posterior","right","inferior"])
medVolReoriented =
medicalVolume with properties:
Voxels: [512×512×264 single]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "unknown"
Orientation: "transverse"
VoxelSpacing: [0.8594 0.8594 1.2453]
NormalVector: [0 0 1]
NumCoronalSlices: 512
NumSagittalSlices: 512
NumTransverseSlices: 264
PlaneMapping: ["coronal" "sagittal" "transverse"]
DataDimensionMeaning: ["posterior" "right" "inferior"]
Modality: "unknown"
WindowCenters: []
WindowWidths: []
To observe the difference in the orientation between volumes, display a transverse slice from each volume in intrinsic coordinates. Display intrinsic coordinates by indexing into the Voxels property.
sliceOriginal = medVol.Voxels(:,:,150); sliceReoriented = medVolReoriented.Voxels(:,:,150); imshowpair(sliceOriginal,sliceReoriented,"montage") title("Original (Left) and Reoriented (Right)")
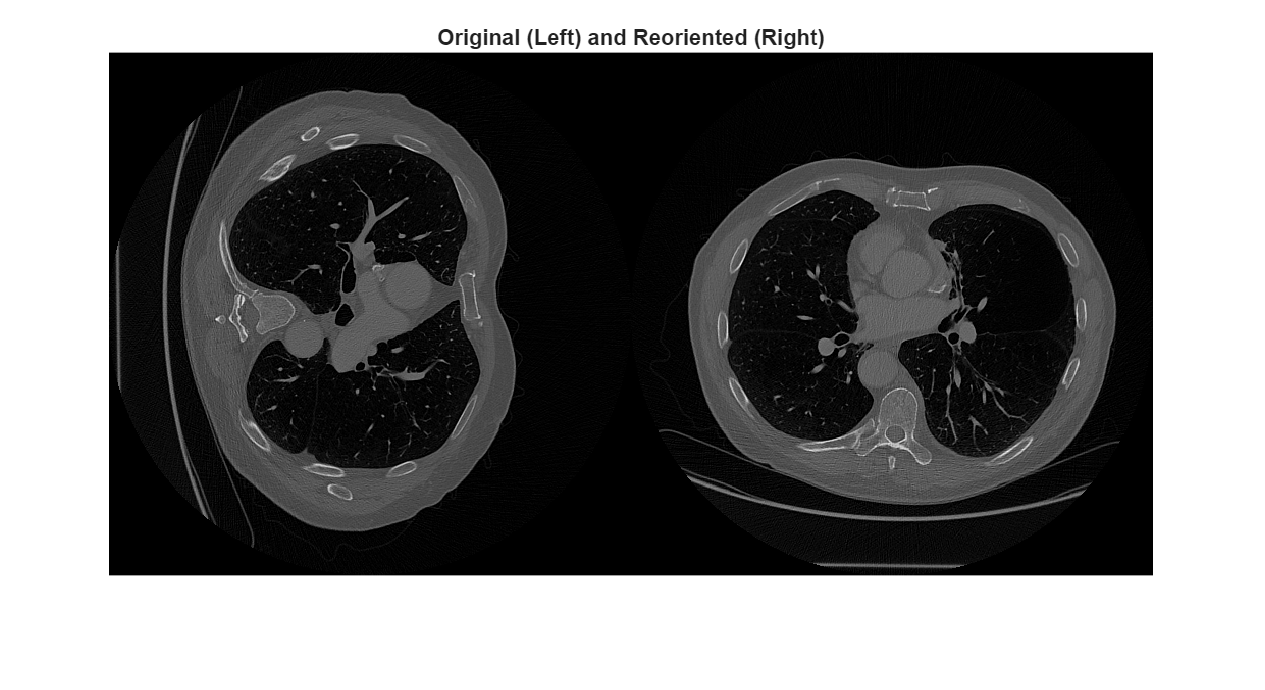
When you display images using functions like imshow and imshowpair, the column indices increase as you move left to right in the display. Therefore, when you look at the patient position within the images, columns increase anteriorly for the original volume slice and to the right for the reoriented volume slice.
[1] Medical Segmentation Decathlon. "Lung." Tasks. Accessed May 10, 2018. http://medicaldecathlon.com/.
The Medical Segmentation Decathlon data set is provided under the CC-BY-SA 4.0 license. All warranties and representations are disclaimed. See the license for details.
Input Arguments
Medical volume, specified as a medicalVolume object.
Primary orientation for the updated volume, specified as one of these options:
| Value | Description |
|---|---|
"coronal" | Change the primary orientation to the coronal plane. Equivalent to specifying
|
"sagittal" | Change the primary orientation to the sagittal plane. Equivalent to specifying
|
"transverse" | Change the primary orientation to the transverse plane. Equivalent to specifying
|
Note
The function reorients the input volume if its DataDimensionMeaning property does not match the equivalent
orientation code for the specified direction. For example, if you
specify direction as "transverse", and the
DataDimensionMeaning value for the input volume is
["anterior","right","inferior"], the function reorients the
volume to ["posterior","left","inferior"] even thought the input
Orientation is already "transverse".
Orientation code for the updated volume, specified as a 3-element string array. The code specifies a new orientation by mapping each anatomical axis to a dimension of the voxel data. The order of the strings determines which data dimension the specified anatomical axis maps to. The content of the strings defines the positive direction for the anatomical axis, as described in the table.
| String | Description |
|---|---|
"left" or "right" | Indicates positive direction of the left/right axis normal to the sagittal plane. |
"inferior" or "superior" | Indicates the positive direction of the inferior/superior axis normal to the transverse plane. |
"posterior" or "anterior" | Indicates the positive direction of the anterior/posterior axis normal to the coronal plane. |
For example, ["anterior","inferior","right"] returns an updated
volume where:
Row indices increase as you move anteriorly through the patient in the coronal plane.
Column indices increase as you move inferiorly through the patient in the transverse plane.
Slice indices increase as you move toward the right side of the patient in the sagittal plane.
Example: ["right","posterior","inferior"]
Example: ["posterior","inferior","right"]
Example: ["superior","anterior","left"]
Data Types: char | string
Output Arguments
Reoriented medical volume, returned as a medicalVolume object. The Voxels property contains the
reoriented voxel data. While dimensions of the voxel data might be permuted and flipped
compared to the original volume, the total number of voxels in the volume remains
unchanged.
Other properties, including VoxelSpacing,
Orientation, PlaneMapping,
DataDimensionMeaning, and NormalVector,
update to reflect the new voxel data. Note that, while the order of the elements of
VoxelSpacing can change, the spacing along each anatomical axis
remains the same. The VolumeGeometry property contains an updated
medicalref3d object that defines the intrinsic-to-world mapping
between the new voxel data and the original position of the volume in the patient
coordinate system.
Tips
The
updateOrientationfunction does not change the orientation or location of the volume in the patient coordinate system. To reposition the volume in patient coordinates, use therepositionfunction.The
updateOrientationfunction permutes and flips the dimensions of the voxel data without changing the spacing or number of slices in each anatomical plane. To resample a medical volume to match the spacing, spatial extents, and orientation of a target volume, use theresampleobject function.
Version History
Introduced in R2025a
See Also
Objects
Functions
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Seleziona un sito web
Seleziona un sito web per visualizzare contenuto tradotto dove disponibile e vedere eventi e offerte locali. In base alla tua area geografica, ti consigliamo di selezionare: .
Puoi anche selezionare un sito web dal seguente elenco:
Come ottenere le migliori prestazioni del sito
Per ottenere le migliori prestazioni del sito, seleziona il sito cinese (in cinese o in inglese). I siti MathWorks per gli altri paesi non sono ottimizzati per essere visitati dalla tua area geografica.
Americhe
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)