resample
Resample medical image volume in different patient coordinate system
Since R2022b
Description
medVolResampled = resample(medVol,R)medicalVolume
object medVol in the patient coordinate system specified by
R. The updated voxel data is returned in a new
medicalVolume object, medVolResampled.
medVolResampled = resample(medVol,R,Name=Value)Method="linear" specifies a linear method for resampling.
Examples
Resample the voxel data of one medical volume using the voxel grid of another volume. This example assumes that the volumes are spatially aligned in the patient coordinate system, but have different voxel spacing, such as CT and PET data acquired on a combined scanner. Resampling achieves one-to-one voxel correspondence between the volumes, which you must have to display the images together as an overlay or perform voxelwise operations such as subtracting the images to calculate a difference map.
Download Data
Download a subset of data from one subject in a data set containing CT and PET images [1][2]. The size of the subset of data is approximately 137 MB. Run this code to download the data set from the MathWorks® website and unzip the folder.
zipFile = matlab.internal.examples.downloadSupportFile("medical", ... "CT-PET-Ventilation-Imaging/Chest-CT-PET.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath)
Specify the path of the folder that contains the downloaded and unzipped data. Each scan consists of a directory of DICOM files.
dataFolder = fullfile(filepath,"Chest-CT-PET");Import Data as medicalVolume Objects
Load the CT and PET volumes as medicalVolume objects. Note that the sizes of the Voxels property, which contains the underlying image, and the values of the VoxelSpacing property, in world units, are different between the volumes.
medVolCT = medicalVolume(fullfile(dataFolder,"3.000000-CT Lung 3.0 B31f-00993"))medVolCT =
medicalVolume with properties:
Voxels: [512×512×175 int16]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "mm"
Orientation: "transverse"
VoxelSpacing: [0.9766 0.9766 2]
NormalVector: [0 0 1]
NumCoronalSlices: 512
NumSagittalSlices: 512
NumTransverseSlices: 175
PlaneMapping: ["sagittal" "coronal" "transverse"]
Modality: "CT"
WindowCenters: []
WindowWidths: []
medVolPET = medicalVolume(fullfile(dataFolder,"4.000000-Galligas Lung-14533"))medVolPET =
medicalVolume with properties:
Voxels: [400×400×159 single]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "mm"
Orientation: "transverse"
VoxelSpacing: [2.0364 2.0364 2.2000]
NormalVector: [0 0 1]
NumCoronalSlices: 400
NumSagittalSlices: 400
NumTransverseSlices: 159
PlaneMapping: ["sagittal" "coronal" "transverse"]
Modality: "PT"
WindowCenters: [159×1 double]
WindowWidths: [159×1 double]
Resample PET Data
Resample the PET volume using the spatial referencing of the CT volume. Verify that the size of the Voxels property and the values of the VoxelSpacing property for the resampled PET volume match those of the CT volume.
petResampled = resample(medVolPET,medVolCT.VolumeGeometry)
Warning: An approximate world to intrinsic mapping is being used.
petResampled =
medicalVolume with properties:
Voxels: [512×512×175 single]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "unknown"
Orientation: "transverse"
VoxelSpacing: [0.9766 0.9766 2]
NormalVector: [0 0 1]
NumCoronalSlices: 512
NumSagittalSlices: 512
NumTransverseSlices: 175
PlaneMapping: ["sagittal" "coronal" "transverse"]
Modality: "unknown"
WindowCenters: []
WindowWidths: []
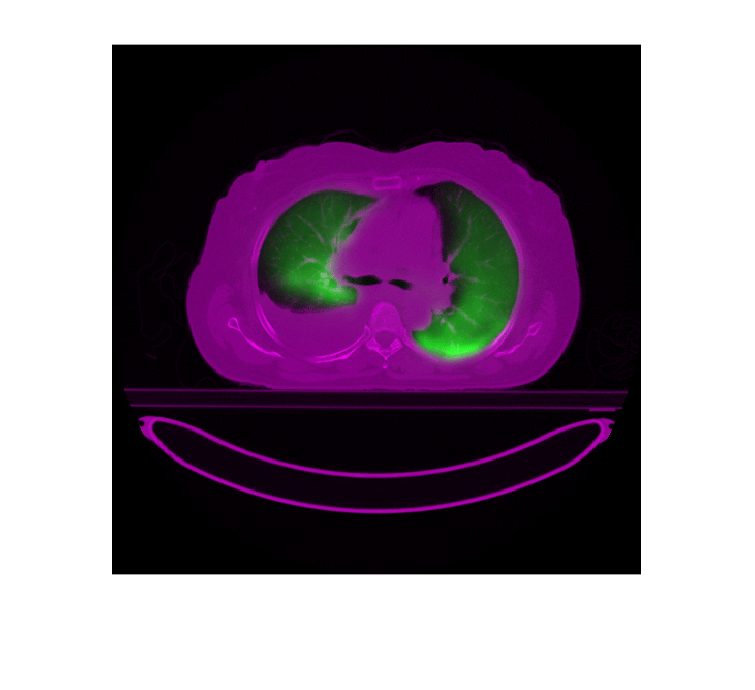
Extract and display a transverse slice from each volume and display them as an overlay.
slicePET = extractSlice(petResampled,90,"transverse"); sliceCT = extractSlice(medVolCT,90,"transverse"); imshowpair(slicePET,sliceCT)
References
[1] Eslick, Enid M., John Kipritidis, Denis Gradinscak, Mark J. Stevens, Dale L. Bailey, Benjamin Harris, Jeremy T. Booth, and Paul J. Keall. "CT Ventilation as a Functional Imaging Modality for Lung Cancer Radiotherapy (CT-vs-PET-Ventilation-Imaging)." The Cancer Imaging Archive, 2022. https://doi.org/10.7937/3PPX-7S22.
[2] Eslick, Enid M., John Kipritidis, Denis Gradinscak, Mark J. Stevens, Dale L. Bailey, Benjamin Harris, Jeremy T. Booth, and Paul J. Keall. "CT Ventilation Imaging Derived from Breath Hold CT Exhibits Good Regional Accuracy with Galligas PET." Radiotherapy and Oncology 127, no. 2 (May 2018): 267–73. https://doi.org/10.1016/j.radonc.2017.12.010.
[3] Clark, Kenneth, Bruce Vendt, Kirk Smith, John Freymann, Justin Kirby, Paul Koppel, Stephen Moore, et al. "The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository." Journal of Digital Imaging 26, no. 6 (December 2013): 1045–57. https://doi.org/10.1007/s10278-013-9622-7.
Apply a geometric transformation to align two volumes in patient coordinates before resampling in the same voxel grid. You must apply a transform when the volumes are not yet aligned in patient coordinates, such as when working with images from different scanners or time points where patient positioning can vary.
Load Images
Download a modified version of the 3-D CT and MRI data sets from The Retrospective Image Registration Evaluation (RIRE) Dataset, provided by Dr. Michael Fitzpatrick. For more information, see the RIRE Project homepage. The modified data set contains one CT scan and one MRI scan from the same patient stored in the NRRD file format. The size of the entire data set is approximately 35 MB. Download the data set from the MathWorks® website, then unzip the folder.
zipFile = matlab.internal.examples.downloadSupportFile("medical", ... "MedicalRegistrationNRRDdataLPS.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath)
Read the CT volume into the workspace as a medicalVolume object.
filenameCT = fullfile(filepath,"supportfilesNRRD","Patient007CT.nrrd"); movingVolume = medicalVolume(filenameCT)
movingVolume =
medicalVolume with properties:
Voxels: [512×512×28 single]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "unknown"
Orientation: "transverse"
VoxelSpacing: [0.6536 0.6536 4]
NormalVector: [0 0 1]
NumCoronalSlices: 512
NumSagittalSlices: 512
NumTransverseSlices: 28
PlaneMapping: ["sagittal" "coronal" "transverse"]
Modality: "unknown"
WindowCenters: []
WindowWidths: []
Read the MRI image into a medicalVolume object. Note that the sizes of the Voxels property, which contains the underlying image, and the values of the VoxelSpacing property, in world units, are different between the volumes.
filenameMRI = fullfile(filepath,"supportfilesNRRD","Patient007MRT1.nrrd"); fixedVolume = medicalVolume(filenameMRI)
fixedVolume =
medicalVolume with properties:
Voxels: [256×256×26 single]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "unknown"
Orientation: "transverse"
VoxelSpacing: [1.2500 1.2500 4]
NormalVector: [0 0 1]
NumCoronalSlices: 256
NumSagittalSlices: 256
NumTransverseSlices: 26
PlaneMapping: ["sagittal" "coronal" "transverse"]
Modality: "unknown"
WindowCenters: []
WindowWidths: []
Transform and Resample Moving Volume
Apply a geometric transformation calculated using the Moment of Mass registration technique in the Medical Registration Estimator app. To learn how to calculate and export the transformation from the app, see Rigid Registration Using Medical Registration Estimator App.
Specify the 4-by-4 transformation matrix from the app. The transformation maps the moving volume to the fixed volume, in the patient coordinate system.
A = [1.0000 0.0041 -0.0004 -6.0548;
-0.0041 1.0000 -0.0020 -19.9711;
0.0042 0.0200 1.0000 -5.4140;
0 0 0 1.0000];Create a geometric transformation object defined by the matrix A.
tform = affinetform3d(A);
Resample the moving volume using the spatial referencing of the fixed volume, specifying the WorldTransform value as the transformation from the registration. Verify that the size of the Voxels property and the value of the VoxelSpacing property for the resampled moving volume match those of the fixed volume.
registeredVolume = resample(movingVolume,fixedVolume.VolumeGeometry, ... WorldTransform=tform, ... FillValue=min(movingVolume.Voxels(:)))
registeredVolume =
medicalVolume with properties:
Voxels: [256×256×26 single]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "unknown"
Orientation: "transverse"
VoxelSpacing: [1.2500 1.2500 4]
NormalVector: [0 0 1]
NumCoronalSlices: 256
NumSagittalSlices: 256
NumTransverseSlices: 26
PlaneMapping: ["sagittal" "coronal" "transverse"]
Modality: "unknown"
WindowCenters: []
WindowWidths: []
Verify that the volumes are aligned in the patient coordinate system.
viewer = viewer3d(BackgroundColor="black",BackgroundGradient="off"); volshow(fixedVolume, ... Parent=viewer, ... RenderingStyle="Isosurface", ... IsosurfaceValue=0.05, ... Colormap=[1 0 1], ... Alphamap=1); volshow(registeredVolume, ... Parent=viewer, ... RenderingStyle="Isosurface", ... IsosurfaceValue=0.05, ... Colormap=[0 1 0], ... Alphamap=1);

Resample a chest CT volume to a target voxel size, without changing the spatial limits in the patient coordinate system. This can be useful to standardize the voxel spacing in a data set of scans acquired using different settings.
The CT volume is saved as a directory of DICOM files. The volume is part of a data set containing three CT scans. The size of the entire data set is approximately 81 MB. Download the data set from the MathWorks® website, then unzip the folder.
zipFile = matlab.internal.examples.downloadSupportFile("medical","MedicalVolumeDICOMData.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath)
Specify the directory of DICOM files for the first CT volume in the data set.
dataFolder = fullfile(filepath,"MedicalVolumeDICOMData","LungCT01");
Create a medical volume object, medVol, for the CT volume. The voxel size is 0.7285-by-0.7825-by-2.5 mm.
medVol = medicalVolume(dataFolder); medVol.VoxelSpacing
ans = 1×3
0.7285 0.7285 2.5000
Resample medVol so that it has a voxel size of 0.5-by-0.5-by-1.25 mm. First, calculate the ratio between the original voxel size and the target voxel size, in millimeters, in each dimension.
targetVoxelSize = [0.5 0.5 1.25]; ratios = targetVoxelSize ./ medVol.VoxelSpacing;
Calculate the target dimensions, in voxels, of the resampled volume.
origSize = size(medVol.Voxels); newSize = round(origSize ./ ratios);
Define the spatial referencing for the resampled volume. First, get the mapping between the intrinsic and patient coordinate systems of medVol by using the intrinsicToWorldMapping object function. The output, origMapping, is an affinetform3d object that contains a transformation matrix in its A property.
origRef = medVol.VolumeGeometry; origMapping = intrinsicToWorldMapping(origRef); tform = origMapping.A;
Transform the matrix tform so that it corresponds to the target voxel size. Create an affinetform3d object, newMapping, that contains the transformed matrix.
newMapping4by4 = tform.* [ratios([2 1 3]) 1]; newMapping = affinetform3d(newMapping4by4);
Create a medicalref3d object that describes the target spatial referencing for the resampled volume.
newRef = medicalref3d(newSize,newMapping);
To maintain the mapping between the patient coordinate axes and the anatomical planes, use the updatePatientCoordinateSystem object function to set the PatientCoordinateSystem property of newRef to match origRef.
newRef = updatePatientCoordinateSystem(newRef,origRef.PatientCoordinateSystem);
Resample medVol by using the resample object function. The voxel size of the new medical volume matches the target size.
newVol = resample(medVol,newRef)
newVol =
medicalVolume with properties:
Voxels: [746×746×176 int16]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "unknown"
Orientation: "transverse"
VoxelSpacing: [0.5000 0.5000 1.2500]
NormalVector: [0 0 1]
NumCoronalSlices: 746
NumSagittalSlices: 746
NumTransverseSlices: 176
PlaneMapping: ["sagittal" "coronal" "transverse"]
DataDimensionMeaning: ["left" "posterior" "superior"]
Modality: "unknown"
WindowCenters: []
WindowWidths: []
Input Arguments
Medical volume, specified as a medicalVolume object.
Spatial referencing object, specified as a medicalref3d
object. R defines the spatial details for the resampled
volume.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: resample(medVol,R,Method="linear") calculates the voxel
values in medVolResampled by linearly interpolating voxel values at the
corresponding locations in medVol.
Fill value, specified as a numeric scalar. The value of
FillValue must be supported by the data type of the voxel data
stored in the Voxels property of R. If an
output voxel in the volume defined by R falls outside the input
volume, medVol, resample sets the
corresponding value of medVolResampled to
FillValue.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Resampling method, specified as "cubic",
"linear", or "nearest".
| Value | Description |
|---|---|
"cubic" | Cubic interpolation. This is the default value, and is suitable for continuous image data, such as CT, MRI, and PET volumes. |
"linear" | Linear interpolation, which is suitable for continuous image data, such as CT, MRI, and PET volumes. |
"nearest" | Nearest neighbor interpolation, which is suitable for discrete data such as binary masks and label images. |
Data Types: char | string
Since R2025a
Geometric transformation to apply before resampling, specified as a geometric
transformation object or as a vector of geometric transformation objects. Use this
argument to apply a transformation that aligns medVol with the
volume defined by R in the patient coordinate system, such as
after registration. By default, the function applies no transformation before
resampling. This table lists the types of geometric transformation objects this
argument accepts.
| Geometric Transformation Object | Description |
|---|---|
transltform3d | Translation transformation |
rigidtform3d | Rigid transformation, consisting of translation and rotation |
simtform3d | Similarity transformation, consisting of translation, rotation, and isotropic scaling |
affinetform3d | Affine transformation, consisting of translation, rotation, anisotropic scaling, reflection, and shearing |
If you specify a vector of geometric transformation objects, the function applies the transformations in the order they appear in the vector.
Output Arguments
Resampled medical volume, returned as a medicalVolume object. The VolumeGeometry property of
medVolResampled contains the spatial referencing information
specified by R. The resampled voxel data is stored in the
Voxels property of medVolResampled.
Tips
To change the location of a medical volume in patient coordinates without resampling the underlying voxel data, use the
repositionfunction.To update the orientation of the voxel data using permutations and flips, without resampling the data or changing the voxel spacing in each anatomical plane, use the
updateOrientationfunction.
Algorithms
The resample function calculates the voxel values in the output
volume, medVolResampled, by mapping locations in the output volume to the
corresponding locations in the input volume (an inverse mapping). When the center of the voxel
in the output volume does not map to the center of a voxel in the input volume,
resample interpolates the input voxel values to calculate the output
values.
resample performs the mapping in the patient coordinate systems of
each volume. The function assumes that the patient coordinate systems of the input and output
volumes have the same origin and orientation convention, specified by the
PatientCoordinateSystem property of a medicalref3d
object.
Version History
Introduced in R2022bApply a geometric transformation to the input volume before resampling by using the new
WorldTransform name-value argument. Use this argument to apply a
transformation that aligns the input medicalVolume object with the volume
defined by R in the patient coordinate system, such as after
registration.
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Seleziona un sito web
Seleziona un sito web per visualizzare contenuto tradotto dove disponibile e vedere eventi e offerte locali. In base alla tua area geografica, ti consigliamo di selezionare: .
Puoi anche selezionare un sito web dal seguente elenco:
Come ottenere le migliori prestazioni del sito
Per ottenere le migliori prestazioni del sito, seleziona il sito cinese (in cinese o in inglese). I siti MathWorks per gli altri paesi non sono ottimizzati per essere visitati dalla tua area geografica.
Americhe
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)