logp
Log unconditional probability density for naive Bayes classifier
Description
lp = logp(Mdl,tbl)lp) of the observations (rows) in tbl using the naive Bayes model Mdl. You can use lp to identify outliers in the training data.
Examples
Compute Unconditional Probability Densities of Observations
Compute the unconditional probability densities of the in-sample observations of a naive Bayes classifier model.
Load the fisheriris data set. Create X as a numeric matrix that contains four petal measurements for 150 irises. Create Y as a cell array of character vectors that contains the corresponding iris species.
load fisheriris
X = meas;
Y = species;Train a naive Bayes classifier using the predictors X and class labels Y. A recommended practice is to specify the class names. fitcnb assumes that each predictor is conditionally and normally distributed.
Mdl = fitcnb(X,Y,'ClassNames',{'setosa','versicolor','virginica'})
Mdl =
ClassificationNaiveBayes
ResponseName: 'Y'
CategoricalPredictors: []
ClassNames: {'setosa' 'versicolor' 'virginica'}
ScoreTransform: 'none'
NumObservations: 150
DistributionNames: {'normal' 'normal' 'normal' 'normal'}
DistributionParameters: {3x4 cell}
Mdl is a trained ClassificationNaiveBayes classifier.
Compute the unconditional probability densities of the in-sample observations.
lp = logp(Mdl,X);
Identify indices of observations that have very small or very large log unconditional probabilities (ind). Display lower (L) and upper (U) thresholds used by the outlier detection method.
[TF,L,U] = isoutlier(lp); L
L = -6.9222
U
U = 3.0323
ind = find(TF)
ind = 4×1
61
118
119
132
Display the values of the outlier unconditional probability densities.
lp(ind)
ans = 4×1
-7.8995
-8.4765
-6.9854
-7.8969
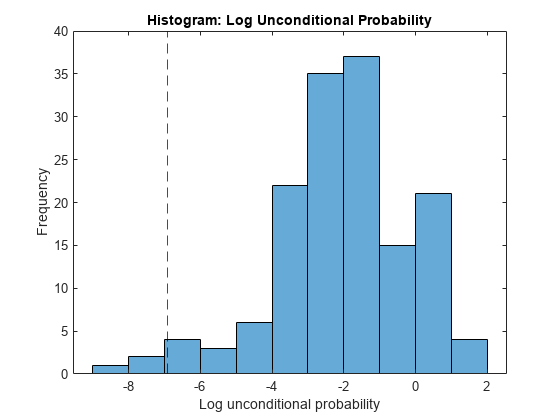
All the outliers are smaller than the lower outlier detection threshold.
Plot the unconditional probability densities.
histogram(lp) hold on xline(L,'k--') hold off xlabel('Log unconditional probability') ylabel('Frequency') title('Histogram: Log Unconditional Probability')
Input Arguments
Mdl — Naive Bayes classification model
ClassificationNaiveBayes model object | CompactClassificationNaiveBayes model object
Naive Bayes classification model, specified as a ClassificationNaiveBayes model object or CompactClassificationNaiveBayes model object returned by fitcnb or compact,
respectively.
tbl — Sample data
table
Sample data used to train the model, specified as a table. Each row of
tbl corresponds to one observation, and each column corresponds
to one predictor variable. tbl must contain all the predictors used
to train Mdl. Multicolumn variables and cell arrays other than cell
arrays of character vectors are not allowed. Optionally, tbl can
contain additional columns for the response variable and observation weights.
If you train Mdl using sample data contained in a table, then the input
data for logp must also be in a table.
X — Predictor data
numeric matrix
Predictor data, specified as a numeric matrix.
Each row of X corresponds to one observation (also known as an
instance or
example), and each column
corresponds to one variable (also known as a
feature). The variables in the
columns of X must be the same as the
variables that trained the Mdl
classifier.
The length of Y and the number of rows of X must
be equal.
Data Types: double | single
More About
Unconditional Probability Density
The unconditional probability density of the predictors is the density's distribution marginalized over the classes.
In other words, the unconditional probability density is
where π(Y = k) is the class prior probability. The conditional distribution of the data given the class (P(X1,..,XP|y = k)) and the class prior probability distributions are training options (that is, you specify them when training the classifier).
Prior Probability
The prior probability of a class is the assumed relative frequency with which observations from that class occur in a population.
Version History
Introduced in R2014b
Comando MATLAB
Hai fatto clic su un collegamento che corrisponde a questo comando MATLAB:
Esegui il comando inserendolo nella finestra di comando MATLAB. I browser web non supportano i comandi MATLAB.

Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list:
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)