densityMap
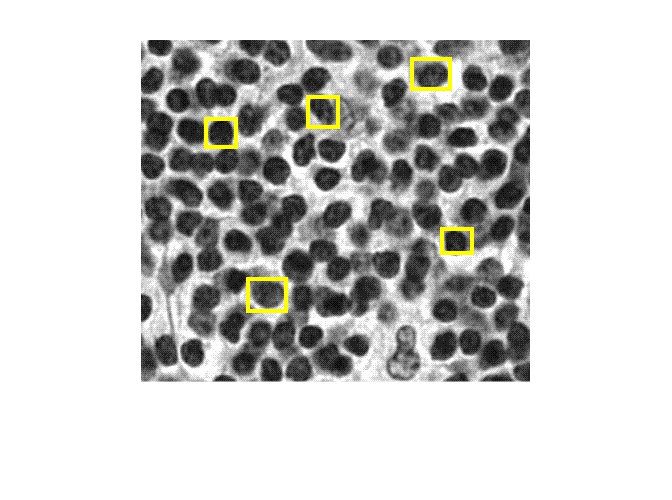
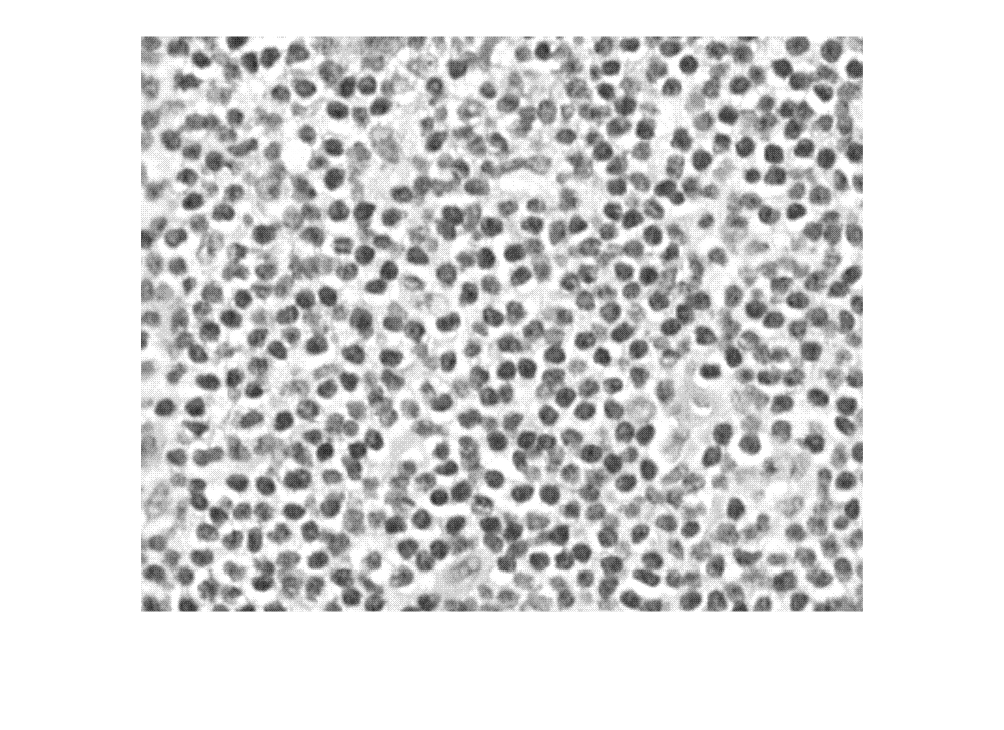
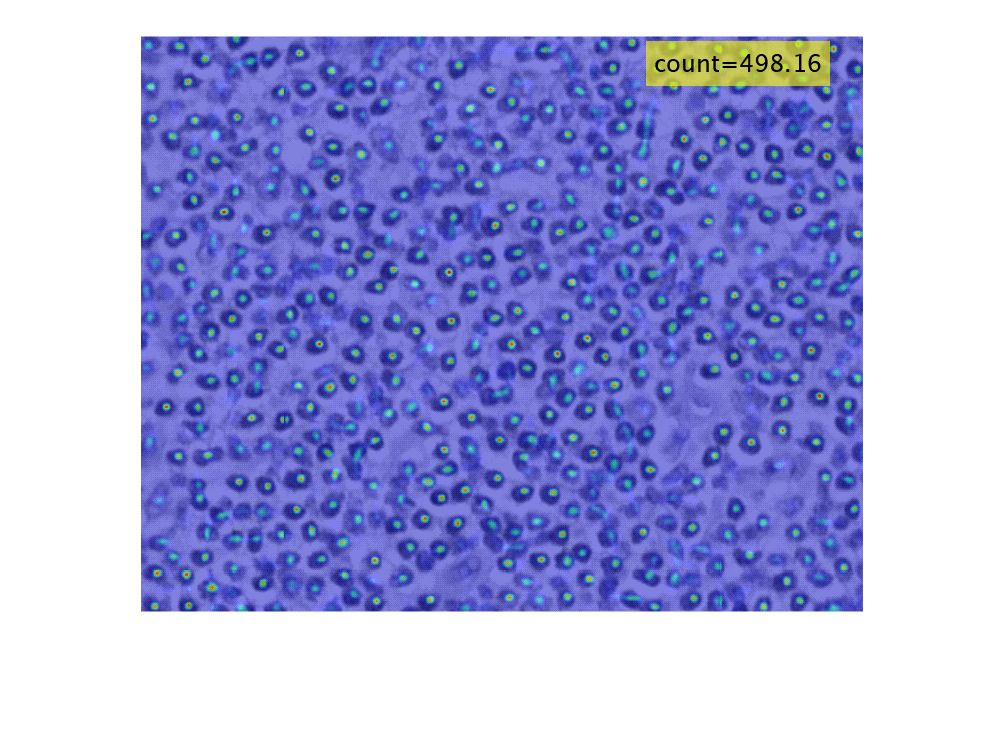
Density map of object count generated using CounTR deep learning model
Since R2025a
Syntax
Description
densMap = densityMap(counter,I)I.
Note
This functionality requires Deep Learning Toolbox™ and the Automated Visual Inspection Library for Computer Vision Toolbox™. You can install the Automated Visual Inspection Library for Computer Vision Toolbox from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons.
densMap = densityMap(___,Name=Value)MiniBatchSize=4
specifies the mini-batch size as 4 when you specify an input datastore
of images, ds.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
References
[1] Liu, Chang, Yujie Zhong, Andrew Zisserman, and Weidi Xie. “CounTR: Transformer-Based Generalised Visual Counting.” arXiv, June 2, 2023. https://doi.org/10.48550/arXiv.2208.13721.
[2] grand-challenge.org. “CAMELYON17 - Grand Challenge.” Accessed November 8, 2024. https://camelyon17.grand-challenge.org/Data/.
Version History
Introduced in R2025a